
Statistical Analysis for Biological Image Data
2025 • Institute of Statistical Science, Academia Sinica
We are an interdisciplinary team at Academia Sinica combining statistics, chemistry, and biology to push the limits of cryo-electron microscopy (cryo-EM). Our work ranges from developing robust algorithms for image analysis and structure classification to pioneering new methods for extracting electrostatic and chemical features directly from cryo-EM maps. By uniting statistical innovation with molecular science, we aim to transform cryo-EM into a tool not only for structural determination but also for quantitative chemical insight that drives discovery in biology and medicine.
Research Highlights (Past 5 Years)
Over the past five years, our group has advanced statistical methodology and structural biology through a series of impactful projects. Each highlight below presents a concise abstract accompanied by a representative figure, showcasing both the scientific problem and our contribution. Together, these works illustrate our continuing efforts to develop robust statistical frameworks for cryo-EM analysis, while also addressing broader challenges in data science and interdisciplinary applications
EM-2SDR: Unsupervised Clustering of 3D Conformations Directly from 2D Cryo-EM Images via Tensor-Structure Modeling
(Annals of Mathematical Sciences and Applications (2025))
We present EM-2SDR, an expectation–maximization algorithm that clusters 3D protein conformations directly from raw 2D cryo-EM particle images. Unlike conventional approaches that reconstruct thousands of 3D volumes before analysis, EM-2SDR integrates a tensor-based dimensionality reduction step within the EM framework, enabling efficient extraction of low-dimensional structural features without intermediate reconstruction. Tests on benchmark ribosome datasets show that EM-2SDR achieves near-perfect classification accuracy, substantially outperforming standard EM-PCA. This approach offers a fast, accurate, and reconstruction-free framework for analyzing structural heterogeneity in large biomolecular complexes.
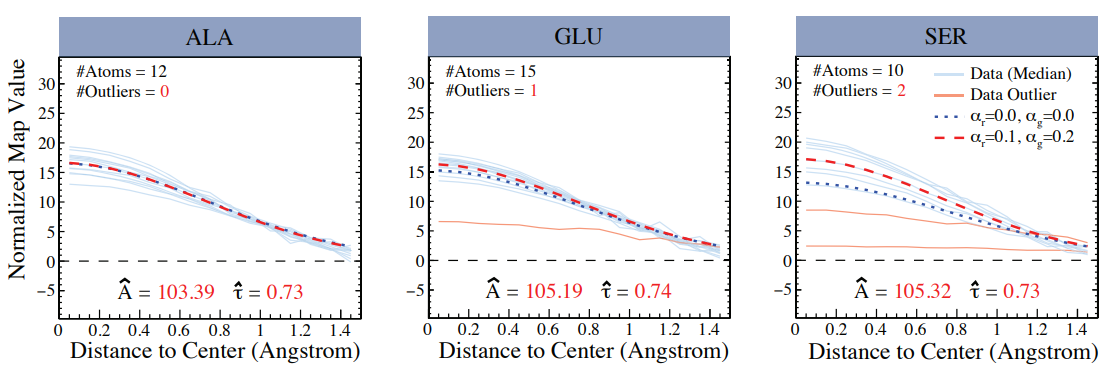
A Hierarchical Robust Linear Model for Cryo-EM Map
Analysis
(bioRxiv (2025))
Cryo-electron microscopy (cryo-EM) has become a pivotal tool for determining the atomic structures of biological macromolecules. In this paper, we propose a hierarchical robust linear (HRL) model to estimate key atom-specific parameters—specifically the amplitude and width of Gaussian functions—that are utilized to assess the consistency between paired cryo-EM maps and their corresponding atomic models, or to improve their refinement. Our HRL modeling leverages minimum density power divergence estimates (MDPDE) to construct a heteroscedastic framework that could potentially mitigate the influence of outliers. We demonstrate the robustness of our method through both simulation studies and real data applications, showing its effectiveness in reducing the impact of outliers and ensuring reliable parameter estimates. Applying our HRL model to analyze real cryo-EM data from human apoferritin (PDB ID: 6Z6U, EMDB ID: 11103) reveals that the Gaussian parameters remain stable across most amino acids, with nitrogen atoms consistently exhibiting lower amplitude and width estimates than expected based on the commonly used Gaussian modeling. These findings highlight the need for a more systematic analysis of paired cryo-EM map and atomic model from the EMDB and PDB to further understand the atom-specific properties in cryo-EM data.
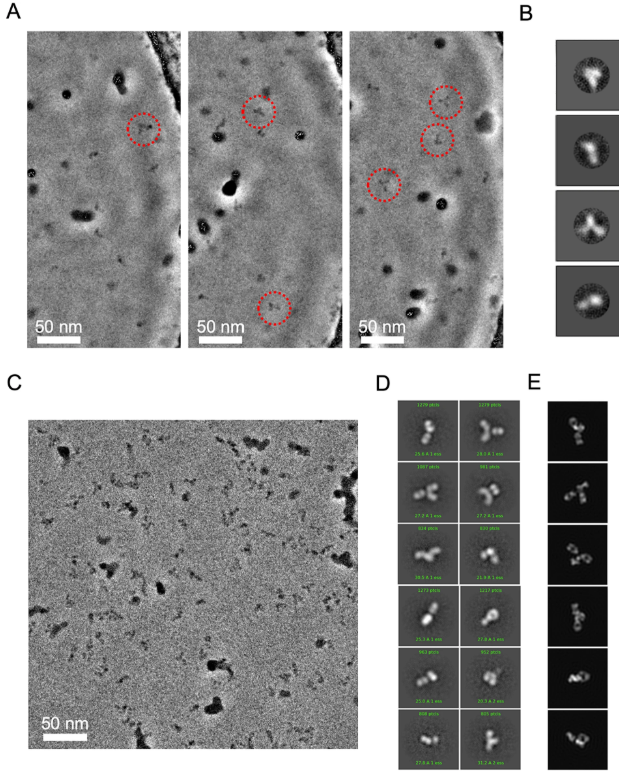
Use of Phase Plate cryo-EM Reveals Conformation Diversity of Therapeutic IgG with 50 kDa Fab Fragment Resolved below
6 Å
(Scientific Reports (2024))
While cryogenic electron microscopy (cryo-EM) is fruitfully used for harvesting high-resolution structures of sizable macromolecules, its application to small or flexible proteins composed of small domains like immunoglobulin (IgG) remain challenging. Here, we applied single particle cryo-EM to Rituximab, a therapeutic IgG mediating anti-tumor toxicity, to explore its solution conformations. We found Rituximab molecules exhibited aggregates in cryo-EM specimens contrary to its solution behavior, and utilized a non-ionic detergent to successfully disperse them as isolated particles amenable to single particle analysis. As the detergent adversely reduced the protein-to-solvent contrast, we employed phase plate contrast to mitigate the impaired protein visibility. Assisted by phase plate imaging, we obtained a canonical three-arm IgG structure with other structures displaying variable arm densities co-existing in solution, affirming high flexibility of arm-connecting linkers. Furthermore, we showed phase plate imaging enables reliable structure determination of Fab to sub-nanometer resolution from ab initio, yielding a characteristic two-lobe structure that could be unambiguously docked with crystal structure. Our findings revealed conformation diversity of IgG and demonstrated phase plate was viable for cryo-EM analysis of small proteins without symmetry. This work helps extend cryo-EM boundaries, providing a valuable imaging and structural analysis framework for macromolecules with similar challenging features.
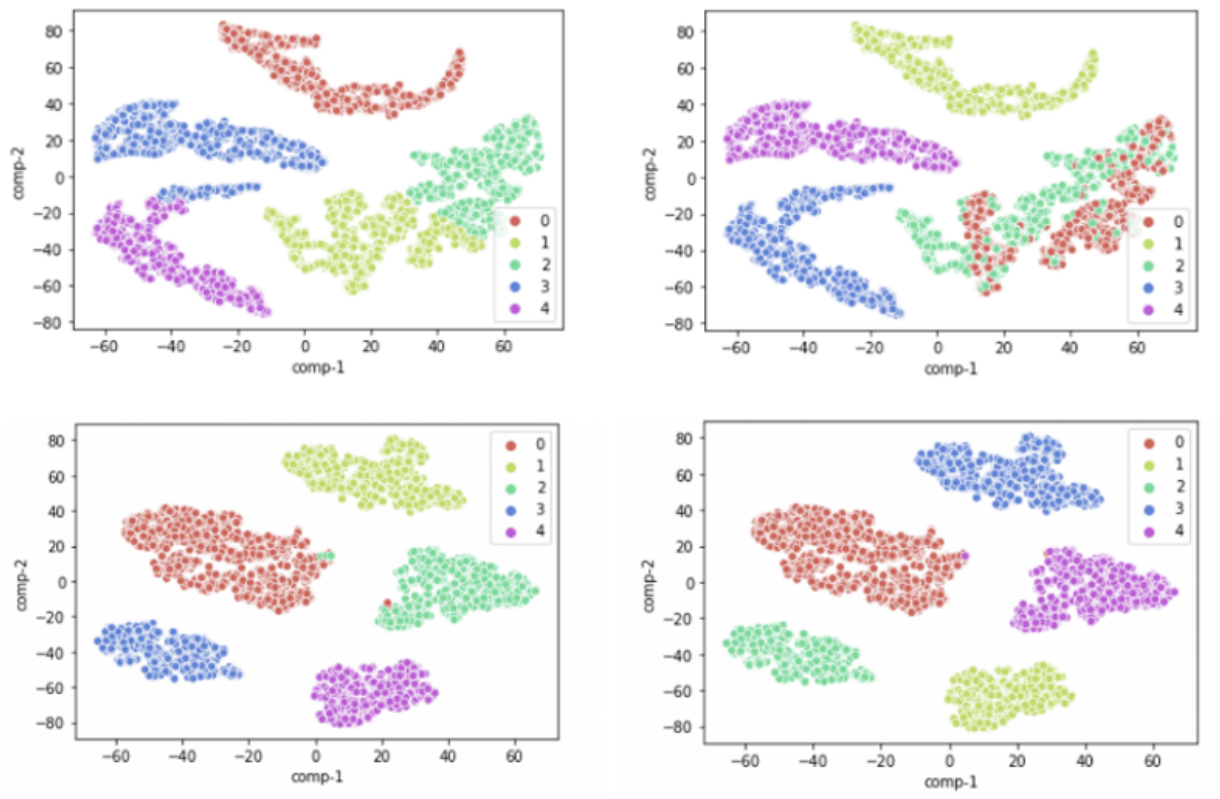
RE2DC: A Robust and Efficient 2D Classifier with Visualization for Processing Massive and Heterogeneous Cryo-EM Data
(bioRxiv(2022))
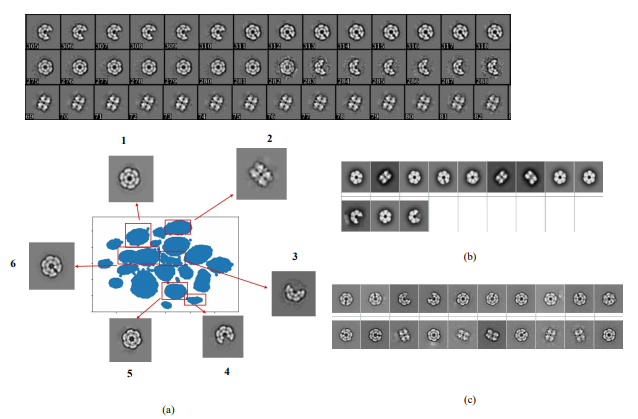
Despite the fact that single particle cryo-EM has become a powerful method of structural biology, processing cryo-EM images are challenging due to the low SNR, high-dimension and un-label nature of the data. Selecting the best subset of particle images relies on 2D classification—a process that involves iterative image alignment and clustering. This process, however, represents a major time sink, particularly when the data is massive or overly heterogeneous. Popular approaches to this process often trade its robustness for efficiency. Here, we introduced a new unsupervised 2D classification method termed RE2DC. It is built upon a highly efficient variant of γ-SUP, a robust statistical cryo-EM clustering algorithm resistant to the attractor effect. To develop this efficient variant, we employed a tree-based approximation to reduce the computation complexity from O(N2) to O(N), with N as the number of images. In addition, we exploited t-SNE visualization to unveil the process of 2D classification. Our tests of RE2DC using various datasets demonstrate it is both robust and efficient, with the potential to reveal subtle structural intermediates. Using RE2DC to curate a dataset of sub-millions of COVID-19 spike particles picked from 3,511 movies only takes 8 hours, suggesting its capability of accelerating cryo-EM structural determination. Currently, RE2DC is available with both CPU and GPU versions, where the implementation only requires modest hardware resources.
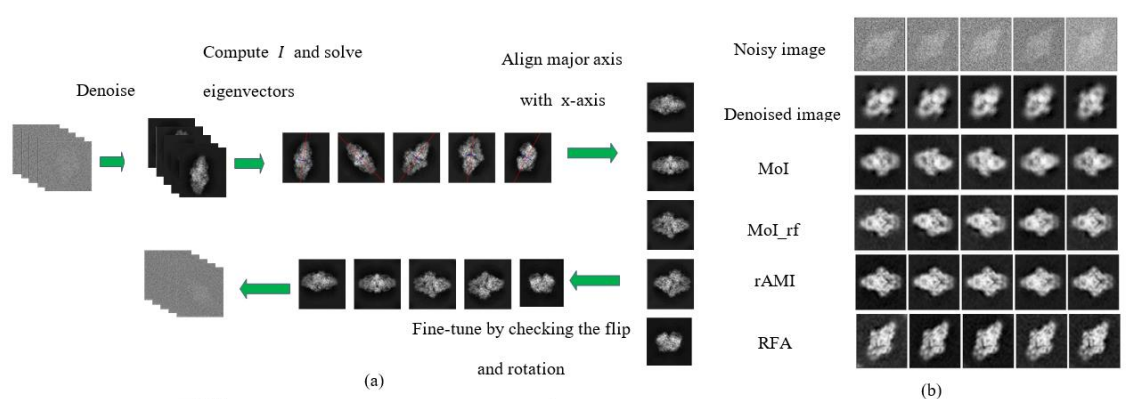
rAMI–Rapid Alignment with Moment of Inertia for Cryo-EM Image Processing
(Microscopy and Microanalysis (2021))
Moment of Inertia (MoI) as a 2 by 2 matrix I containing the central moments with order two, whose first eigenvector corresponds to the object’s orientation, has been a popular tool for image alignment (Jan, Suk and Zitová, 2016). However, the low SNR nature of cryo-EM images has prevented the direct application of MoI. We proposed an algorithm called Rapid Alignment with Moment of Inertia (rAMI), and we show that it can be widely applicable to the current alignment steps in cryo-EM.
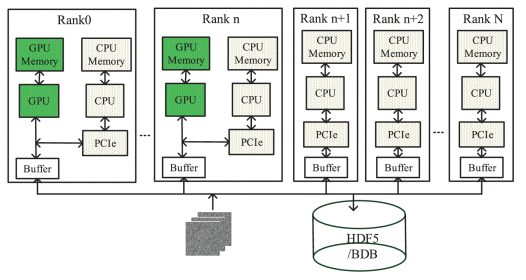
Cryo-Ralib - A Modular Library for Accelerating Alignment in CRYO-EM
(2021 IEEE International Conference on Image Process (ICIP) (2021))
Thanks to GPU-accelerated processing, cryo-EM has become a rapid structure determination method that permits capture of dynamical structures of molecules in solution, which has been recently demonstrated by the determination of COVID-19 spike protein in March, shortly after its breakout in late January 2020. This rapidity is critical for vaccine development in response to the emerging pandemic. Compared to the Bayesian-based 2D classification widely used in the work-flow, the multi-reference alignment (MRA) is less popular. It is time-consuming despite its superior in differentiating structural variations. Interestingly, the Bayesian approach has higher complexity than MRA. We thereby reason that the popularity of Bayesian is gained through GPU acceleration, where a modular acceleration library for MRA is lacking. Here, we introduce a library called Cryo-RALib that expands the functionality of CUDA library used by GPU ISAC. It contains a GPU-accelerated MRA routine for accelerating MRA-based classification algorithms. In addition, we connect the cryo-EM image analysis with the python data science stack to make it easier for users to perform data analysis and visualization. Benchmarking on the TaiWan Computing Cloud (TWCC) shows that our implementation can accelerate the computation by one order of magnitude.
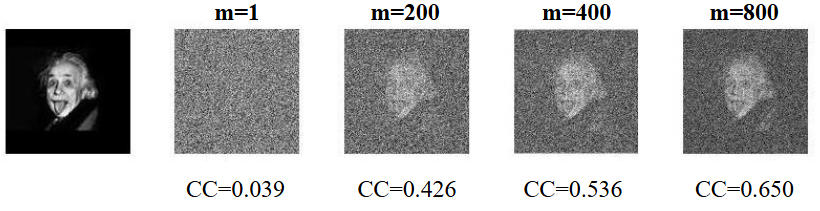
Quantification of model bias underlying the phenomenon of Einstein from Noise
(Statistica Sinica (2021))
“Einstein from Noise” states a pitfall in cryo-electron microscopy analysis that a processing output could be heavily biased towards the imposed model. We develop a simple mathematical framework under which an image is expressed as a vector of dimension p where p is large and show how the bias is formed by averaging a properly chosen set of purely noise images that are most highly correlated with the target image.
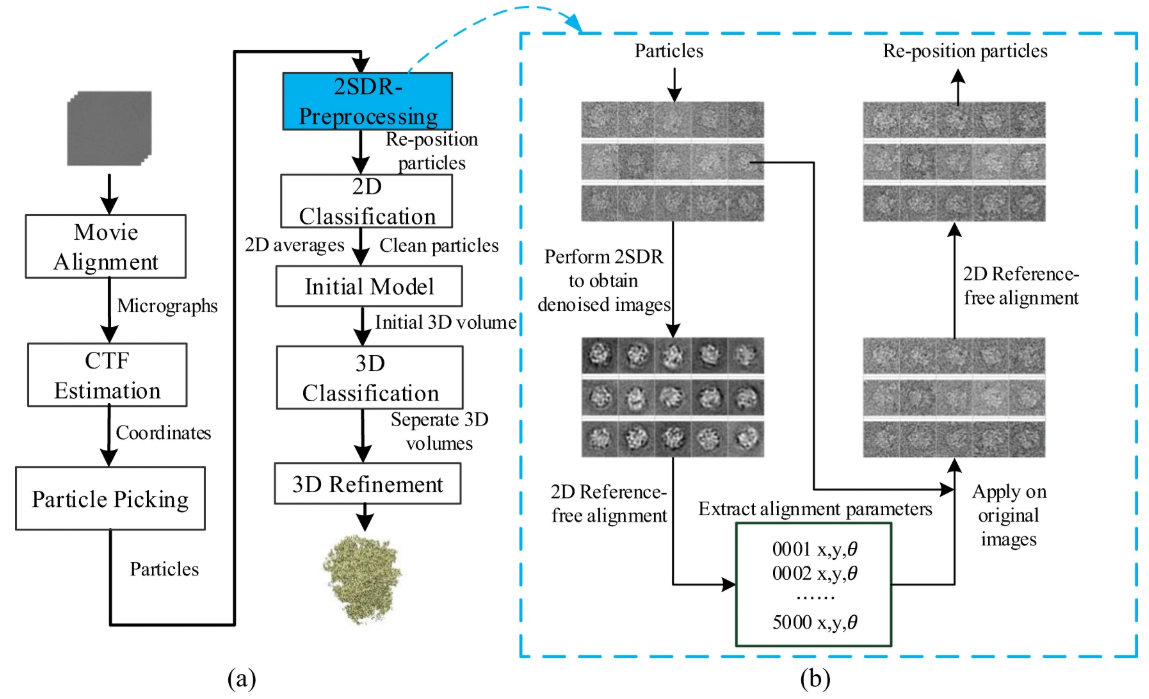
Pre-Pro: a Fast Pre-Processor for Single-Particle Cryo-EMthrough Enhancing 2D Classification
(Communications Biology (2020))
2D classification plays a pivotal role in analyzing single particle cryo-electron microscopy images. Itis mainly used to curate cryo-EM images by harvesting good particle datasets by separating good par-ticles from bad ones or non-particle contaminants. Due largely to the presence of heavy noise, theclassification results are often non-ideal while the computation burden is further aggravated by recentexpansion in size and number of images. Here, we introduce a simple and loss-less pre-processor thatincorporates a fast dimension-reduction (2SDR) de-noiser to enhance 2D classification. By implement-ing this 2SDR pre-processor prior to representative classification algorithms including RELION andISAC, we compare the performances with and without the pre-processor. Tests on multiple cryo-EMexperimental datasets show the pre-processor can make the classification faster, improve the yield ofparticles, and increase the number of good classes to generate better initial models. Testing on thenanodisc-embedded TRPV1 dataset with high heterogeneity with a 3D reconstruction workflow usingthe initial model generated by class-averages, we found that the pre-processor improved the resolutionto 2.82 Å toward the Nyquist frequency. Those findings and analyses suggest the 2SDR de-noiser, ofminimal cost, is widely applicable for boosting the performances of 2D classification algorithms. Thegeneralization of the pre-processing strategy to accommodate neural network de-noisers is discussed.
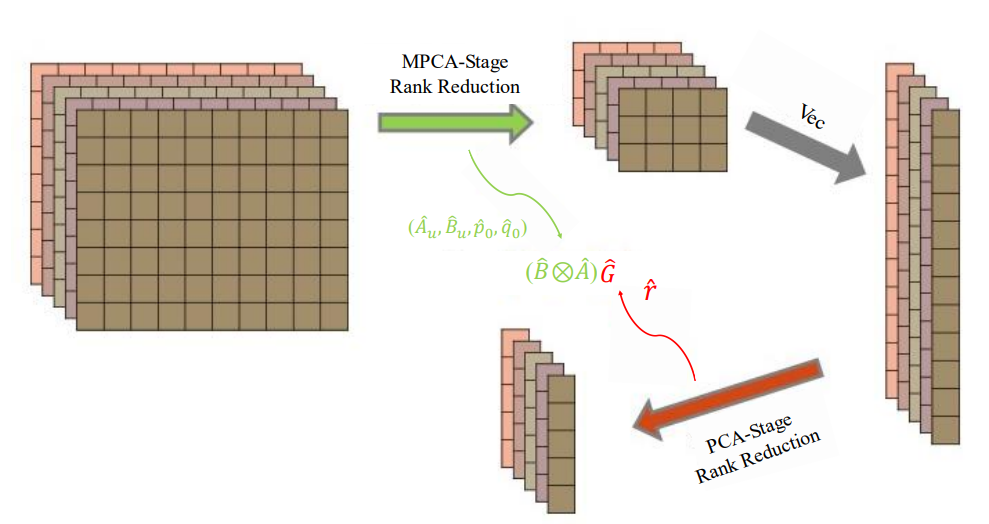
Two-stage dimension reduction for noisy high-dimensional images and application to Cryogenic Electron Microscopy
(Annals of Mathematical Sciences and Applications (2020))
Principal component analysis (PCA) is arguably the most widely used dimension-reduction method for vector-type data. When applied to a sample of images, PCA requires vectorization of the image data, which in turn entails solving an eigenvalue problem for the sample covariance matrix. We propose herein a two-stage dimension reduction (2SDR) method for image reconstruction from high-dimensional noisy image data. The first stage treats the image as a matrix, which is a tensor of order 2, and uses multilinear principal component analysis (MPCA) for matrix rank reduction and image denoising. The second stage vectorizes the reduced-rank matrix and achieves further dimension and noise reduction. Simulation studies demonstrate excellent performance of 2SDR, for which we also develop an asymptotic theory that establishes consistency of its rank selection. Applications to cryo-EM (cryogenic electronic microscopy), which has revolutionized structural biology, organic and medical chemistry, cellular and molecular physiology in the past decade, are also provided and illustrated with benchmark cryo-EM datasets. Connections to other contemporaneous developments in image reconstruction and high-dimensional statistical inference are also discussed.
Collaboration Works
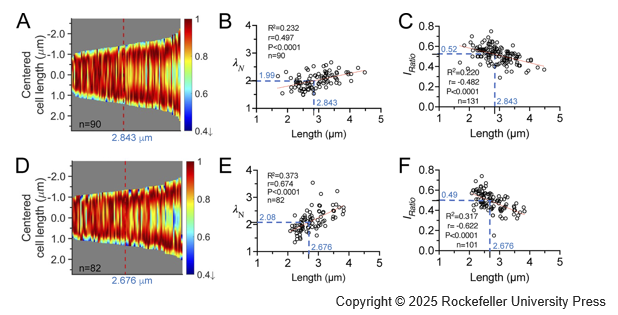
Growth-Dependent Concentration Gradient of the Oscillating Min System in Escherichia Coli
(Journal of Cell Biology (2025))
Cell division in Escherichia coli is intricately regulated by the MinD and MinE proteins, which form oscillatory waves between cell poles. These waves manifest as concentration gradients that reduce MinC inhibition at the cell center, thereby influencing division site placement. This study explores the plasticity of the MinD gradients resulting from the interdependent interplay between molecular interactions and diffusion in the system. Through live cell imaging, we observed that as cells elongate, the gradient steepens, the midcell concentration decreases, and the oscillation period stabilizes. A one-dimensional model investigates kinetic rate constants representing various molecular interactions, effectively recapitulating our experimental findings. The model reveals the nonlinear dynamics of the system and a dynamic equilibrium among these constants, which underlie variable concentration gradients in growing cells. This study enhances quantitative understanding of MinD oscillations within the cellular environment. Furthermore, it emphasizes the fundamental role of concentration gradients in cellular processes.
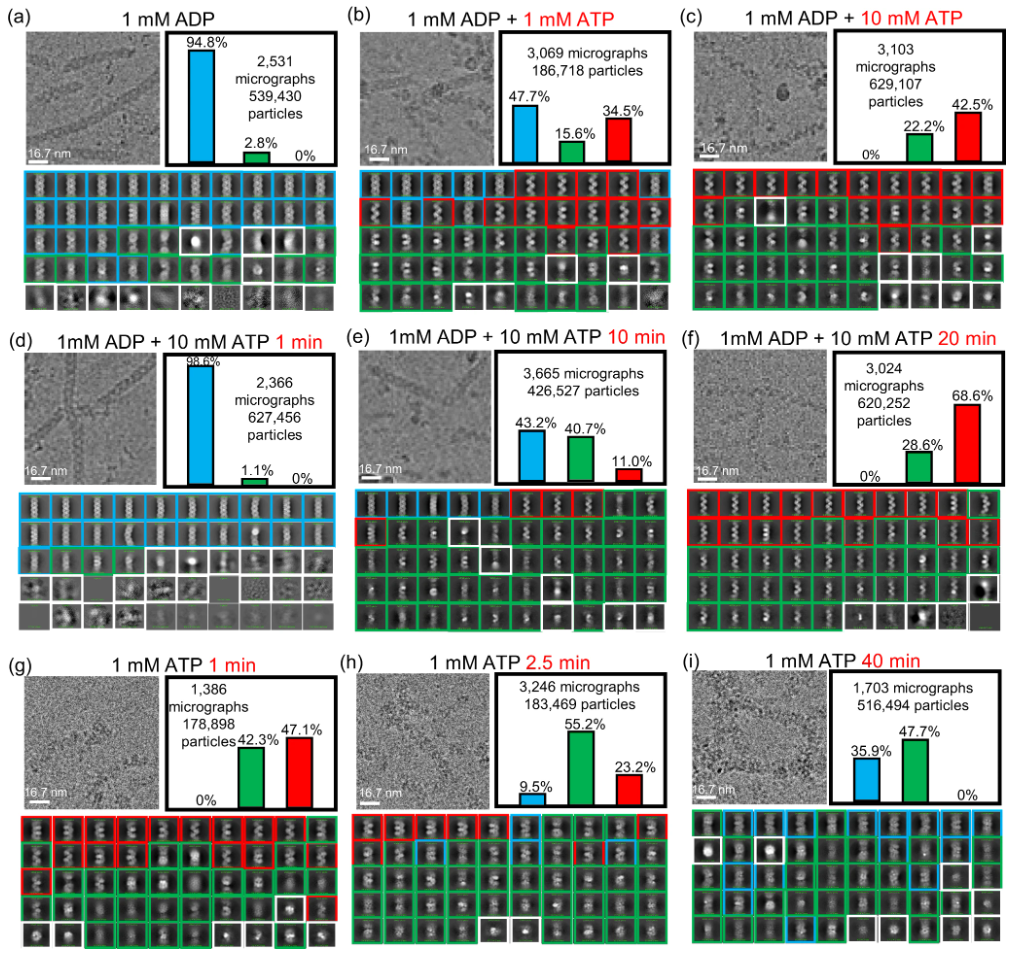
A RAD51–ADP Double Filament Structure Unveils the Mechanism of Filament Dynamics in Homologous Recombination
(Nature Communications (2024))
ATP-dependent RAD51 recombinases play an essential role in eukaryotic homologous recombination by catalyzing a four-step process: 1) formation of a RAD51 single-filament assembly on ssDNA in the presence of ATP, 2) complementary DNA strand-exchange, 3) ATP hydrolysis transforming the RAD51 filament into an ADP-bound disassembly-competent state, and 4) RAD51 disassembly to provide access for DNA repairing enzymes. Of these steps, filament dynamics between the ATP- and ADP-bound states, and the RAD51 disassembly mechanism, are poorly understood due to the lack of near-atomic-resolution information of the ADP-bound RAD51–DNA filament structure. We report the cryo-EM structure of ADP-bound RAD51–DNA filaments at 3.1 Å resolution, revealing a unique RAD51 double-filament that wraps around ssDNA. Structural analysis, supported by ATP-chase and time-resolved cryo-EM experiments, reveals a collapsing mechanism involving two four-protomer movements along ssDNA for mechanical transition between RAD51 single- and double-filament without RAD51 dissociation. This mechanism enables elastic change of RAD51 filament length during structural transitions between ATP- and ADP-states.
More Publications
▪
Deriving a Sub-Nanomolar Affinity Peptide From TAP to Enable smFRET Analysis of RNA Polymerase II Complexes
(Methods (2019))
▪
The generalized degrees of freedom of multilinear principal component analysis
(Journal of Multivariate Analysis (2019))